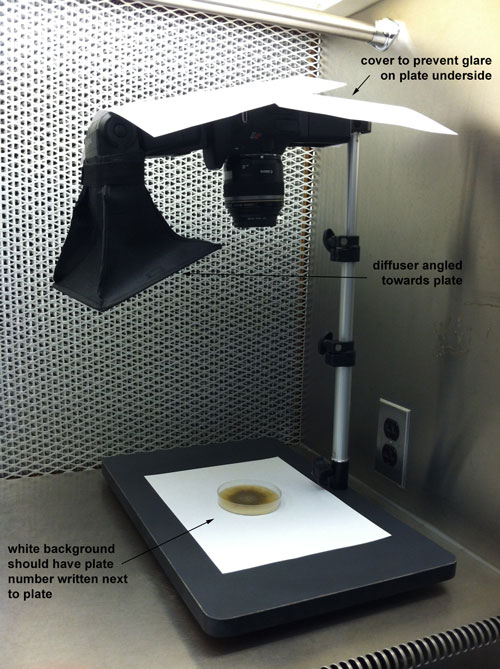
These are the conditions on our USDA permit for hand-carrying live fungi through airport customs. All fungi have to be on minislants.
1. At least TWENTY days prior to each hand carry incident, the permit holder or designee must notify the PPQ Permit Compliance Officer by email (
re*********************@ap***.gov
) to provide specific information on the hand carrier’s identity, the anticipated first port of arrival into the United States, the actual date of arrival, the time, and, if travel is by airline, the flight number. The Compliance Officer will notify Customs and Border Protection (CBP) Agriculture Specialist at the port of entry to document and facilitate the entry of the organisms.
2.The hand carrier must indicate that living organisms are being imported under a USDA permit on the Customs Declaration form if such form is required at the port of entry.
3.At the port of entry, individuals carrying permitted organism must also present to CBP officers the following articles: Passport or Visa and a valid hand carry PPQ Form 599 Red/White label corresponding to the permit.
4.Inspection by CBP Officers must confirm that all hand carried articles are securely packaged as per the permit conditions. In the event that a problem is detected, the CBP officer may seize the package and require its movement to the nearest PPQ Inspection Station for processing, clearance or destruction. The permit holder will be responsible for all costs incidental to such forwarding.
5.After CBP confirmation and clearance through the first port of entry into the United States, hand carried organisms must be transported directly to the containment facility authorized in the permit.
6.Upon arrival at the facility, the PPQ Compliance Officer must be notified within 24 hours that the organisms arrived. Notification may be by fax (301-734-5392) or email (
re*********************@ap***.gov
). Notification must be by an independent third party (e.g. containment facility director, departmental chair, campus biosafety officer, etc.). The notification must include the permit number, label number, date of arrival, the specific organisms that were imported, their origin, and quantity. Failure to notify the PPQ compliance officer may result in loss of hand carry privileges. A PPQ inspector may also visit the facility to confirm the arrival of the package and its contents.
7. ONLY PERSON(S) WHOSE NAME(S) IS/ARE LISTED IN THE ISSUED PERMIT IS/ARE AUTHORIZED TO HAND CARRY.
8. You will receive NEW PPQ Form 599 Red/White labels for each hand carry event once you have submitted the required information. You can NOT use the red and white labels described above that are prepared for BONDED CARRIERS. If you use the PPQ Form 599 Red/White labels for bonded carrier while attempting to hand carry, the package will be seized by the Department of Homeland Security and destroyed.
9. An authorization to hand-carry includes only the organism identified in the permit. Presence of unauthorized organisms in any packages on an individual authorized to hand-carry is a permit violation. Presence of unauthorized organism at the receiving containment facility at any time is also evidence of a permit violation.